
Hire The Best Metagenomics Tutor
Top Tutors, Top Grades. Without The Stress!
10,000+ Happy Students From Various Universities
Choose MEB. Choose Peace Of Mind!
How Much For Private 1:1 Tutoring & Hw Help?
Private 1:1 Tutors Cost $20 – 35 per hour* on average. HW Help cost depends mostly on the effort**.
Metagenomics Online Tutoring & Homework Help
What is Metagenomics?
Metagenomics is the study of genetic material from environmental samples using high-throughput molecular methods. Instead of isolating individual microbes, it directly analyzes DNA from soil, water, or the human gut. Next Generation Sequencing (NGS) platforms enable the profiling of microbial communities, uncovering species diversity and metabolic potential in complex ecosystems.
Alternative names include environmental genomics, community genomics, ecogenomics and microbiome shotgun sequencing.
Key topics span sample collection and DNA extraction from various habitats (for example collecting soil at a contaminated site for bioremediation studies), library preparation and sequencing technology selection (such as choosing Illumina or Nanopore platforms), data preprocessing including quality control and read trimming, assembly of fragmentary sequences into contigs, taxonomic assignment using reference databases, functional annotation to predict gene roles, comparative metagenomics across different environments, statistical analysis of diversity metrics, and visualization of complex datasets. Functional metagenomics further explores gene function through cloning and heterologous expression, often revealing novel enzymes for industrial or medical applications.
In 1998, Norman Pace and colleagues coinced the term metagenomics while studying ribosomal RNA sequences in environmental samples. A breakthrough came in 2004 when Craig Venter’s Global Ocean Sampling expedition applied shotgun sequencing to marine microbes, vastly expanding known diversity. By 2005, human gut microbiome studies emerged, linking community shifts to health and disease. The following years saw development of robust bioinformatics pipelines for taxonomic and functional analysis. In 2010 the Earth Microbiome Project was launched to catalog microbial diversity globally. Recent advances integrate multi-omics approaches, combining metatranscriptomics and metaproteomics to capture functional dynamics.
How can MEB help you with Metagenomics?
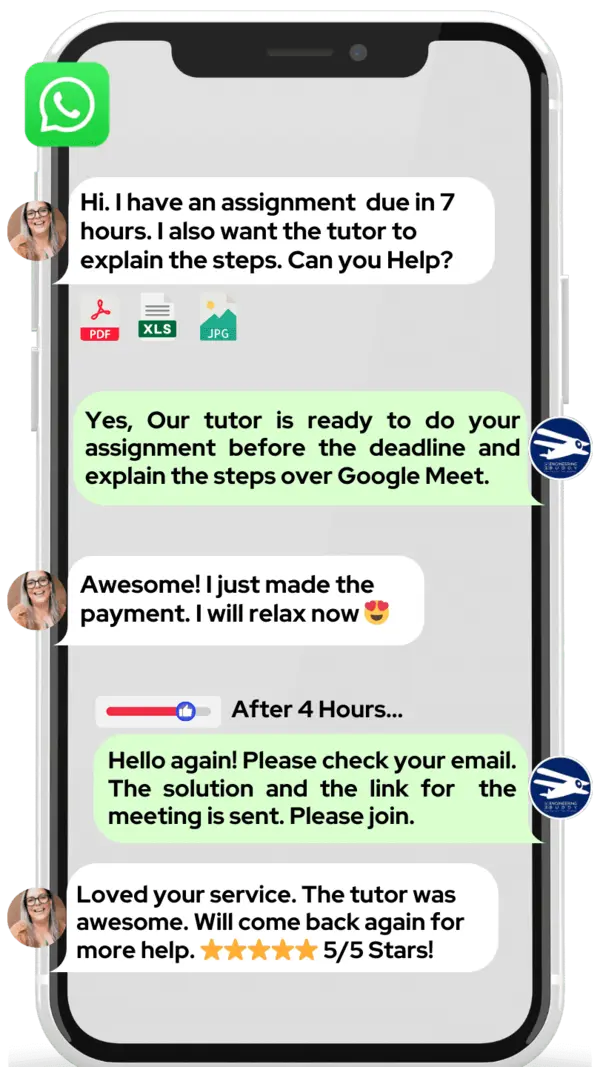
Do you want to learn metagenomics? At MEB, we offer private one‑on‑one online metagenomics tutoring. If you are a school, college, or university student and want top grades on assignments, lab reports, live assessments, projects, essays, or dissertations, try our 24/7 instant online metagenomics homework help. We prefer WhatsApp chat, but if you don’t use it, email us at meb@myengineeringbuddy.com.
Most of our students come from the USA, Canada, the UK, the Gulf, Europe, and Australia.
Students ask us for help when a subject is hard, there are too many assignments, topics are confusing, or solving problems takes too long. They also reach out when they have health or personal issues, part‑time jobs, missed classes, or can’t keep up with their professor’s pace.
If you are a parent and your ward is struggling with this subject, contact us today. Help your ward ace exams and homework—they will thank you!
MEB also supports more than 1,000 other subjects with expert tutors. Getting help when you need it makes learning easier and school less stressful.
DISCLAIMER: OUR SERVICES AIM TO PROVIDE PERSONALIZED ACADEMIC GUIDANCE, HELPING STUDENTS UNDERSTAND CONCEPTS AND IMPROVE SKILLS. MATERIALS PROVIDED ARE FOR REFERENCE AND LEARNING PURPOSES ONLY. MISUSING THEM FOR ACADEMIC DISHONESTY OR VIOLATIONS OF INTEGRITY POLICIES IS STRONGLY DISCOURAGED. READ OUR HONOR CODE AND ACADEMIC INTEGRITY POLICY TO CURB DISHONEST BEHAVIOUR.
What is so special about Metagenomics?
Metagenomics stands out because it allows students to study entire microbial communities directly from soil, water or human gut samples, without growing each microbe in a lab. This subject uncovers the full range of DNA from bacteria, viruses and fungi, revealing unknown species. By focusing on environmental DNA, metagenomics gives a complete picture of microbial diversity that traditional microbiology misses.
Compared to other biomedical courses, metagenomics offers high‑throughput sequencing and big data analysis skills, giving students practical bioinformatics experience. It helps in disease research and environmental studies but demands strong computing knowledge and can be costly. Unlike focused subjects, it covers broad microbial ecosystems and complex data interpretation, which delivers valuable insights but requires more time and resources to master.
What are the career opportunities in Metagenomics?
Many universities now offer master’s and PhD programs in metagenomics or microbiome science. Students can also take short courses in bioinformatics and data analysis, often online, to stay current with new sequencing tools. Workshops on long‑read sequencing and AI‑driven methods are especially popular today.
Common job roles include bioinformatics analyst, research scientist, data specialist, and clinical microbiome consultant. Analysts build pipelines to process DNA data from soil, water, or human samples. Scientists design experiments to track how microbes affect health, crops, or pollution cleanup.
We learn metagenomics to understand whole microbial communities without growing them in a lab dish. This helps in early disease detection, tracking new pathogens, and studying gut health. Test prep and coursework build skills in DNA sequencing, statistics, and software like QIIME or Mothur.
Metagenomics is used in medicine for personalized treatments, in agriculture to boost plant growth, and in environment to monitor pollution. Its main advantages are speed, the ability to find rare or unknown microbes, and insights into how microbial networks work together.
How to learn Metagenomics?
Start with a solid foundation in molecular biology and statistics. Learn basic genetics concepts, then pick up command‑line skills in Linux. Next, study data analysis tools like Python or R, and explore key metagenomic pipelines (QIIME, mothur, Galaxy). Download public datasets from NCBI and practice running simple workflows. Work through one tutorial at a time, build small projects, and review your results. Repeat until you can handle raw sequence files, run analyses, and interpret diversity metrics.
Metagenomics brings together biology, computing, and statistics, so it can feel tough at first. You might struggle with data formats, command lines, or interpreting community profiles. But if you break it into small steps—one tool, one concept at a time—it quickly becomes manageable. Many learners find that regular practice and clear tutorials turn hard topics into routine tasks.
You can definitely start metagenomics on your own using free courses, videos, and practice data. But a tutor can speed up your learning, help you avoid common mistakes, and answer questions right away. If you run into roadblocks with code errors, file formats, or data interpretation, an experienced guide saves you hours of frustration and gets you back on track.
Our MEB tutors are experts in metagenomics and bioinformatics. We offer 24/7 one‑on‑one online sessions, personalized study plans, hands‑on project support, and assignment help. Whether you need to master a pipeline, debug your scripts, or prep for an exam, we tailor each lesson to your pace and goals. Flexible schedules and affordable rates mean you get expert help when you need it most.
Time to reach basic proficiency varies by background. If you already know genetics and some coding, expect about 3–4 months of part‑time study (5–10 hours/week). Complete beginners might need 5–6 months. Intensive study (15–20 hours/week) can cut that to 6–8 weeks. Always factor in extra time for hands‑on practice and project work.
Check YouTube channels like EMBL‑EBI Metagenomics, Biostars tutorials and Next Gen Seq Reads. Visit NCBI’s Metagenomics portal, Coursera’s “Metagenomics” course, Khan Academy for stats and molecular bio basics, and Galaxy Project training pages. Key books include “Metagenomics for Microbiology” by Kostas Kostidis, “Metagenomics: Current Innovations and Future Trends” (ed. Robert W. Li), “Bioinformatics Data Skills” by Vince Buffalo, and “Handbook of Molecular Microbial Ecology” by Kenneth Timmis. Many students also use GitHub repos for practice datasets and scripts.
College students, parents, tutors from USA, Canada, UK, Gulf etc. if you need a helping hand, be it online 1:1 24/7 tutoring or assignments, our tutors at MEB can help at an affordable fee.








